LPS Extraction Kit
Products capable of separating / purifying LPS present in the outer wall of gram-negative bacteria
- LPS isolation / purification present in gram negative bacteria
- Phenol-water type LPS extraction
- Effective removal of contaminants such as proteins and nucleic acids
- The most efficient and easy to extract LPS
- The entire process of extracting LPS is completed within a maximum of 60 minutes
Código: 17141

Sin existencias
199,00€
Sin existencias
Description
Products capable of separating / purifying LPS present in the outer wall of gram-negative bacteria
- LPS isolation / purification present in gram negative bacteria
- Phenol-water type LPS extraction
- Effective removal of contaminants such as proteins and nucleic acids
- The most efficient and easy to extract LPS
- The entire process of extracting LPS is completed within a maximum of 60 minutes
LPS Extraction Kit
Is a product that can separate and purify LPS present in the outer wall of Gram-negative bacteria. In general, Gram-negative bacteria have a separate structure called lipopolysaccharide (LPS) in addition to phospholipids and proteins in the external layer of the cell envelope. These LPSs play an important role in bacterial growth and survival, and are known to play many roles, especially in the interaction between host and parasite.
In addition, LPS is understood to be closely related to the immune response, such as causing peripheral vascular collapse by secretion of endotoxin when septic shock is applied, and is also important for toxic pathophysiological reactions. This kind of LPS has a different extraction pattern for each bacterium, which allows phylogenetic separation of bacteria. As mentioned earlier, bacterial endotoxin is usually present on the outer wall of Gram-negative bacterium cells.
It is not expressed during the life of the bacteria, but when the bacteria grow or die, the cell wall is broken and released to the outside. Natural entotoxin consists of LPS, Proteins, and Phospholipids. It is very stable in its natural state, strong in heat, negatively charged, and has a large molecular weight (over 1,000,000 dalton). Generally, proteins are extracted with Trichloroacetic acid, Butanol or EDTA to remove lipid associated proteins (LAP), and phospholipids are extracted using phenol.
Typically, when you try to extract LPS, you can get about 1-4% LPS per bacterial dry weight. Since the LPS Extraction Kit is based on the Phenol water method, the protein in entotoxin is denatured to denature it as a precipitate between the water layer and the phenol layer, and the lipid is separated by the phenol / chloroform fraction.
Kit Contents
| Contents | 17141(100 Rxn) |
|---|---|
| Lysis Buffer | 100 ml x 1 ea |
| Purification Buffer | 80 ml x 1 ea |
Technical Data
LPS extraction yield
| Strain | LPS yield (μg) | Protein contamination* |
|---|---|---|
| S. typhimurium | 200 ~ 400 | < 0.2 μg |
| S. enteritidis | 90 ~ 250 | < 0.2 μg |
| S. gallinarum | 150 ~ 450 | < 0.2 μg |
| E. coli (wild type) | 220 ~ 490 | < 0.2 μg |
| E. coli (O:055) | 260 ~ 510 | < 0.2 μg |
| E. coli (O:111) | 220 ~ 500 | < 0.2 μg |
| E. coli (O:1) | 180 ~ 380 | < 0.2 μg |
| E. coli (O:2) | 180 ~ 380 | < 0.2 μg |
5 LPS was extracted from each G (-) strain of OD600 using the LPS Extraction Kit, and the extracted LPS was quantified using the Purpald Assay. As a result, the above extraction efficiency was confirmed. On the other hand, the presence of protein was observed using our SMART ™ Micro BCA Assay Kit and the degree of contamination was confirmed to be less than 0.2 μg.
Band pattern is given by extracted LPS from various species
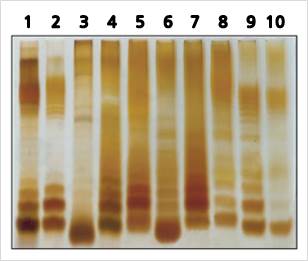
Lane 1 : S. typhimurium Lane 2 : S. enteritidis
Lane 3 : E. coli (O055) Lane 4 : E. coli (O111)
Lane 5 : S. gallinarium Lane 6 : S. enterititis
Lane 7 : S. typhimurium Lane 8 : E. coli (wild tyoe)
Lane 9 : E, coli (O111) Lane 10 : E. coli (O2)
As a result of silver staining after SDS-PAGE of LPS extracted from various kinds of gram-negative bacteria, a ladder type band pattern was observed depending on the characteristics of LPS present in a multimeric form and the number of multimers, Patterns are observed to be different.




















